 |
Software & Hardware
Software
HaarSeg: Fast and Flexible Microarray Segmentation
Erez Ben-Yaacov and Yonina C. Eldar
Introduction
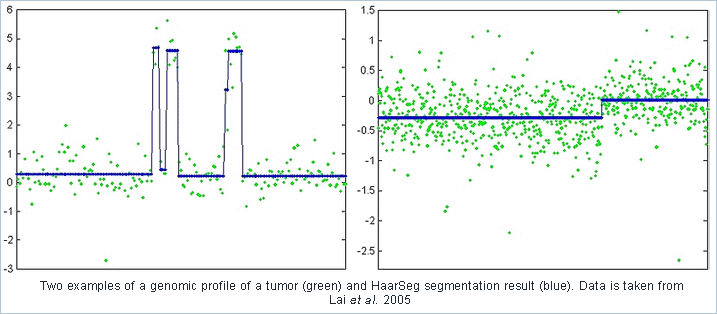
A central task in the analysis of aCGH and Tiling microarray data is the segmentation into groups of probes sharing the same copy number. Some well known segmentation methods suffer from very long running times, preventing interactive data analysis.
We suggest a new 1-D piecewise constant segmentation method, based on wavelet decomposition and thresholding, which detects significant breakpoints in the data. Our algorithm is over 1,000 times faster than leading approaches, with similar performance. Another key advantage of the proposed method is its simplicity and flexibility. Due to its intuitive structure it can be easily generalized to incorporate several types of side information. We consider two extensions which include side information indicating the reliability of each measurement, and compensating for a changing variability in the measurement noise. The resulting algorithm outperforms existing methods, both in terms of speed and performance, when applied to real high density aCGH data.
Reference
Software Download
Installation:
1. Unzip all files to a directory of your choice.
2. Compile the mex functions: (compiled windows 32-bit versions are provided).
(-) In your matlab environment, set the directory to where you unzipped the sources.
(-) Type the following in the matlab environment:
>> mex mexConvAndPeak.c
>> mex mexThresAndUnify.c
>> mex mexAdjustBreaks.c
Usage:
1. HaarSeg.m is the main function, used to segment data. Type "help HaarSeg.m" for basic usage instructions.
2. thresBySig.m is the function of the aberration threshold, which can be applied on the segmentation result.
Type "help thresBySig.m" for basic usage instructions.
Usage:
1. HaarSeg.R is the main function, used to segment data. See comments inside HaarSeg.R for usage instructions
|
|

